Downloading cloud-free satellite image time series using Python (the efficient way)
For a recent project, I wanted to analyse satellite images from the European Space Agency’s Sentinel-2 mission over specific areas and over time. The Sentinel-2 mission is pretty great: it has 10-meter resolution optical imagery, takes a picture of almost all points on the continental earth every five days, and is completely free to access. My areas of interest were quite small (low single-digit square kilometres), whereas Sentinel-2 satellite images are usually published in huge 100x100km2 chunks. This post describes how I went about this in what seems to me a fairly efficient way. All data and software used here is open access/open source.
We take a two-step process to process as little unnecessary data as possible and keep our analysis relatively fast. First, we identify those images that might potentially be relevant for us by looking at image metadata only (see section 2.). Second, we look at the actual image data in our area of interest, check for clouds, and download only if it is actually useful for us (see section 3.). The analysis uses Sentinel-2 images in COG format hosted on AWS S3.
1. Define Areas of Interest
First, we need to define the areas that we are interested in. Here, I picked a number of parks in Berlin. For each park we define a name and a polygon describing the location and shape of the park in Well-Known-Text (WKT) format. I used this web-based tool to draw polygons on a map around my areas of interest and export them as WKT.
We also import a bunch of packages here - the important ones are described in the following sections.
import os
from typing import Tuple
import geopandas as gpd
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
import numpy as np
import pandas as pd
import pyproj
from pystac_client import Client
import rasterio
from rasterio.mask import mask
from rasterio.plot import show
import shapely
from shapely.geometry import LinearRing, Polygon
from shapely.ops import transform
parks = [
(
"Tiergarten",
"POLYGON ((13.351501 52.513962, 13.360061 52.514732, 13.370124 52.511389, 13.363086 52.510175, 13.352037 52.510018, 13.351501 52.513962))",
),
(
"Humboldthain",
"POLYGON ((13.381429 52.544103, 13.383102 52.542851, 13.389539 52.545043, 13.387608 52.547914, 13.384089 52.547679, 13.381858 52.5464, 13.383102 52.545461, 13.381429 52.544103))",
),
(
"VolksparkFriedrichshain",
"POLYGON ((13.427434 52.527997, 13.437046 52.529068, 13.438548 52.528624, 13.44211 52.53053, 13.445027 52.528076, 13.440951 52.526979, 13.439192 52.527841, 13.435158 52.526483, 13.437819 52.524055, 13.434128 52.523663, 13.427434 52.527997))",
),
(
"TreptowerPark",
"POLYGON ((13.460611 52.490908, 13.462799 52.493155, 13.472153 52.491143, 13.478933 52.484662, 13.473441 52.482101, 13.460611 52.490908))",
),
(
"TempelhoferFeld",
"POLYGON ((13.395079 52.479696, 13.405463 52.479226, 13.415418 52.475671, 13.415847 52.470443, 13.405034 52.467096, 13.394993 52.469606, 13.390874 52.475043, 13.395079 52.479696))",
),
(
"Hasenheide",
"POLYGON ((13.41027 52.487746, 13.409412 52.484714, 13.410828 52.484113, 13.410528 52.482414, 13.418809 52.480585, 13.420182 52.481108, 13.420054 52.486857, 13.41027 52.487746))",
),
(
"Grunewald",
"POLYGON ((13.189399 52.434874, 13.250846 52.483617, 13.208623 52.497414, 13.193518 52.489052, 13.197294 52.483198, 13.193518 52.477344, 13.195235 52.472534, 13.200727 52.471279, 13.196608 52.46354, 13.191459 52.462285, 13.190772 52.454963, 13.191115 52.448059, 13.18116 52.44764, 13.181504 52.439269, 13.189399 52.434874))",
),
(
"GaertenDerWelt",
"POLYGON ((13.571628 52.540188, 13.579952 52.53917, 13.580467 52.537317, 13.576863 52.536351, 13.574932 52.534655, 13.568753 52.53502, 13.571628 52.540188))",
),
(
"ParkAmGleisdreieck",
"POLYGON ((13.374437 52.496512, 13.377054 52.496146, 13.376282 52.495193, 13.377569 52.494945, 13.37905 52.496891, 13.378664 52.49403, 13.376282 52.49339, 13.375789 52.49207, 13.3736 52.492345, 13.373514 52.494579, 13.374437 52.496512))",
),
]2. Identifying Suitable Satellite Images Based on STAC Metadata
SENTINEL-2 data can be accessed in a number of ways. Here, I use the catalogue hosted on AWS S3 here in the wonderful COG format (see next section). This data is searchable through a STAC API. The STAC or SpatioTemporal Asset Catalog specification provides a unified way to query our data and allows us to filter the many available images with spatial, temporal, or other metadata filters. In Python, we can use the pystac-client library to identify images that seem relevant to us. We just point it to the catalogue on S3 and we are ready to query.
An image that is useful to our analysis should fulfil three properties. First, it should overlap one of the areas of interest we defined in the previous step. Second, it should be taken in the timeframe that we are interested in. Here, we are searching for images taken in 2022. Finally, it should not contain clouds over the areas of interest.
Whereas filtering by area and time is straightforward, the question of cloud cover is more difficult. In the metadata we can use to filter the Sentinel-2 STAC, we have an attribute eo:cloud_cover that gives us the percentage of clouds in the whole image. However, remember that the Sentinel-2 images are 100km x 100km in size. This limits the usefulness of the average cloud cover value: an average cloud-cover of 70% could still leave our area of interest cloud-free, or conversely the clouds covering 4% of a different image could all be concentrated over our area of interest. An example of such a scenario is given in the image below. Based on the cloud-cover metadata alone, we cannot know with certainty whether an image is useful to us or not.
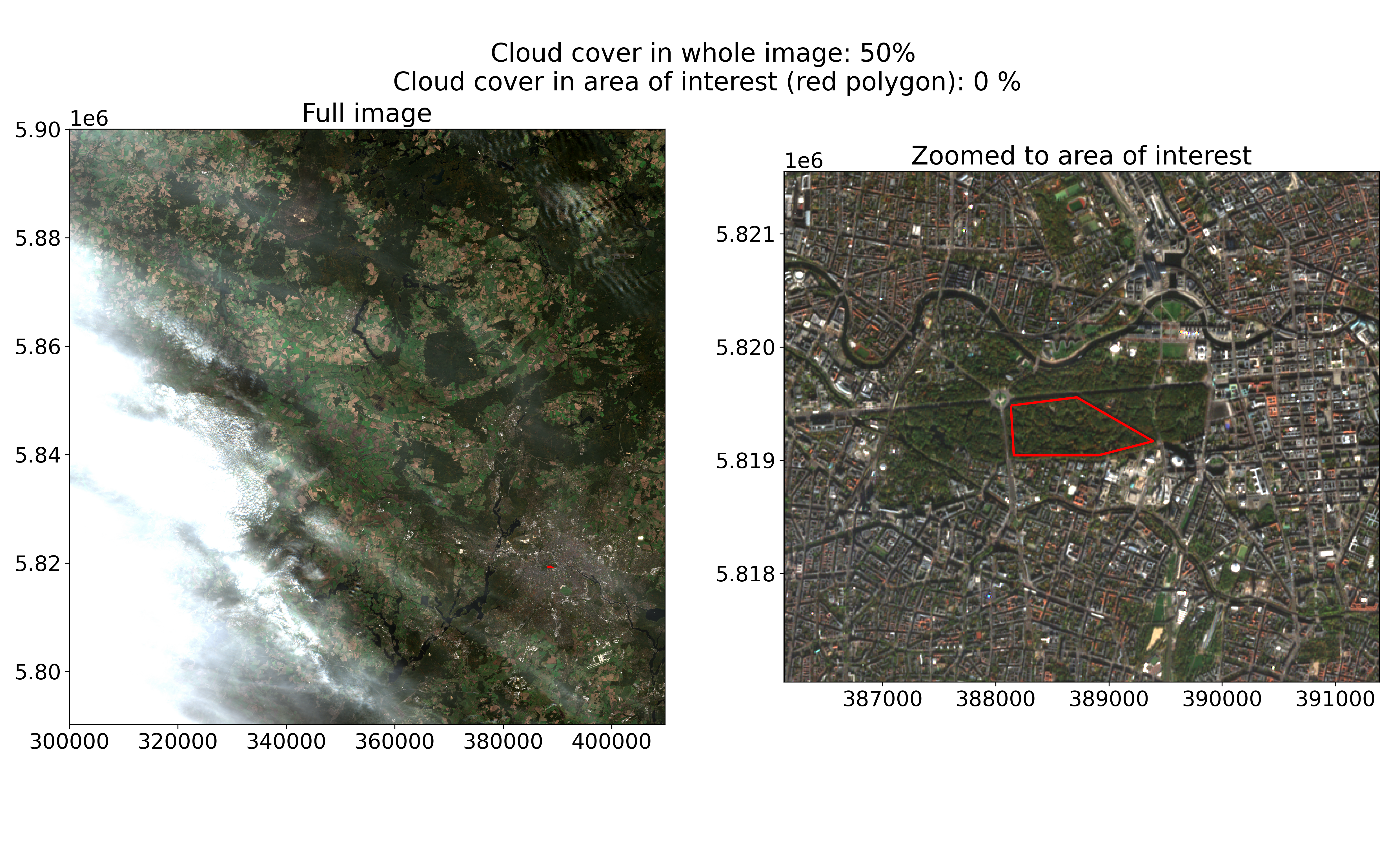
Sentinel-2 image from 2022-11-06 showing clouds in the west, but not over Berlin’s Tiergarten park.
To illustrate this further, I analysed all 272 images Sentinel-2 took in 2022 of Berlin’s Tiergarten park and plotted the fraction of clouds in the whole 100x100km image versus the park area only (following the methodology described in the next section). As the park is quite small, we can see that in most cases it is either fully covered in clouds or fully free. We also see that there were days where the cloud cover in the whole image is as high as 80%, but our area of interest was completely uncovered. You can find the code used to generate this plot on my GitHub at the end of the notebook here.
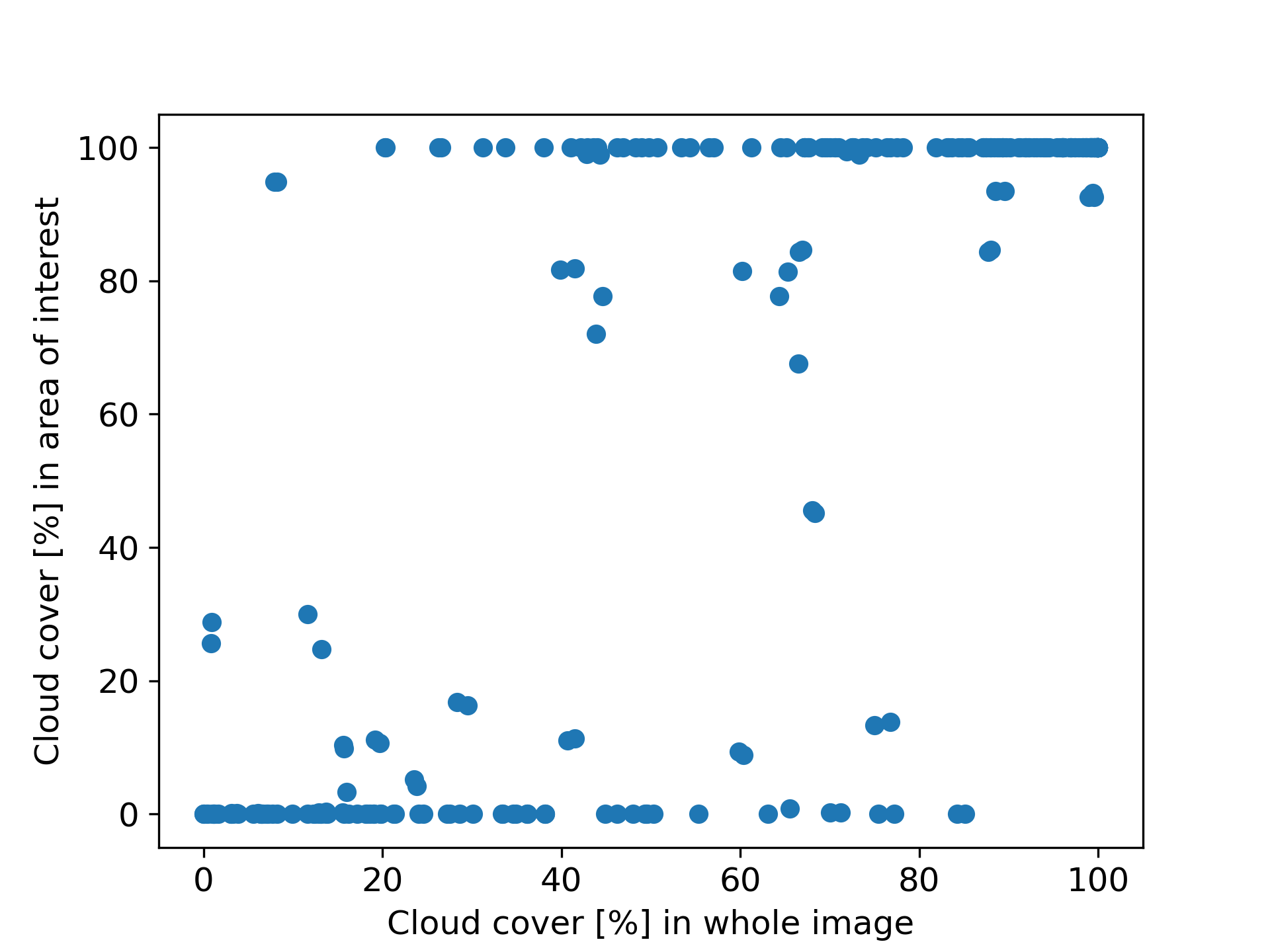
Cloud cover in the whole image versus our area of interest for 272 Sentinel-2 images containing Berlin’s Tiergarten park.
We consequently address the cloud problem in two steps. In this first querying step we filter by a leniently high cloud cover percentage to discard the images that are unlikely to contain useful data to us (e.g. 80%). In the next step (section 3.) we then check precisely whether there are clouds over our area of interest. Here, we apply a much stricter filter (e.g. 2%).
At the end of this step, we have identified 1034 images in the year 2022 with less than 50% cloud cover that we want to examine in greater detail in the next step. The median park in our collection has 109 images. Note that so far, we did not download any imagery, but just queried metadata.
# search parameters
DATE_RANGE = "2022-01-01/2022-12-31"
# if an image has less than <MAX_CLOUD_COVER_SEARCH> % of clouds
# overall, we add it to our search and check the precise
# cloud cover over our area of interest in the next step
MAX_CLOUD_COVER_SEARCH = 80
def execute_sentinel2_query(catalog: Client, bounding_box: tuple,
date_range: str, max_cloud_cover: int
) -> gpd.GeoDataFrame:
"""find sentinel 2 TCI images with max_cloud_cover
in date_range that overlap bounding_box.
Returns DF with metadata of relevant images"""
query = catalog.search(
collections=["sentinel-2-l2a"], datetime=date_range,
limit=200, bbox=bounding_box,
query={"eo:cloud_cover":{"lt":max_cloud_cover}},
)
stac_json = query.item_collection_as_dict()
return gpd.GeoDataFrame.from_features(stac_json, "epsg:4326")
catalog = Client.open("https://earth-search.aws.element84.com/v1")
image_metadata_df = []
for (area_name, area_wkt) in parks:
area_geom = shapely.from_wkt(area_wkt)
area_bbox = area_geom.bounds
images_over_area = execute_sentinel2_query(catalog, area_bbox,
DATE_RANGE, MAX_CLOUD_COVER_SEARCH)
images_over_area["area_name"] = area_name
images_over_area["area_geom"] = area_geom
image_metadata_df.append(images_over_area)
image_metadata_df = pd.concat(image_metadata_df)
print(f"Total number of images: {len(image_metadata_df)}")
print(f"Median number of images per area:",
image_metadata_df.groupby("area_name").size().median(), "\n")
image_metadata_df[["area_name", "s2:granule_id", "datetime",
"eo:cloud_cover", "earthsearch:s3_path"]].sample(3).head()3. Checking if Clouds Cover Our Area of Interest And Downloading the Relevant Color Image Sections
Now that we have the list of images that fulfil our query criteria, for each image we need to check whether there are clouds over our areas of interest to see whether it is relevant for us.
As we are only interested in a small area within a huge image, it would be unfortunate if we had to download the whole image everytime to then only use a few hundred pixels within it. Luckily, we can take a more efficient route as the data is available on AWS in Cloud-Optimized Geotiff or COG format. Every COG file is a GeoTIFF file, essentially meaning that pixel values map to points on earth. The useful part about COGs is that a file organizes its pixel values in a spatially regular way and contains information about this regular layout in its header. Provided with our area of interest, software designed for handling COGs (such as GDAL which we use through Rasterio) can thus read the header, use the information to compute where within the huge file the pixels that interest us are located, and use a HTTP range request to download only those pixels. This drastically reduces the amount of data we need to download.
To check for clouds, we use the Scene Classification Layer (SCL) produced by ESA as part of the Level-2A processing algorithm for each image. This is published as a COG file, where each pixel indicates which of eleven classes it likely contains, including water, vegetation, and different types of cloud. You can read more about this here.
The main advantage of this approach is its simplicity, but anecdotally the performance of the SCL cloud detection algorithm seems to vary quite a bit. I will leave a proper exploration of this for a future post. For our current purposes, however, we continue with this simple approach.
Using Rasterio, we download the part of the scene classification COG over our area of interest and determine the fraction of pixels that contain clouds or cloud shadows. If this is over our threshold (e.g. 2%), we discard the image. If the image is sufficiently free of clouds, we download the true colour image that overlaps our area of interest and save it locally.
# if the area of interest has more than MAX_CLOUD_COVER_PRECISE % cloud
# cover, we do not download it
MAX_CLOUD_COVER_PRECISE = 2
def project_shape(shape: Polygon, from_proj_epsg: int,
to_proj_epsg: int) -> Polygon:
"""Projects a shapely Polygon to a new CRS"""
from_crs = pyproj.CRS('EPSG:'+str(from_proj_epsg))
to_crs = pyproj.CRS('EPSG:'+str(to_proj_epsg))
project = pyproj.Transformer.from_crs(
from_crs, to_crs, always_xy=True).transform
return transform(project, shape)
def s3path_to_tif_download_link(s3path:str, image_type: str):
""" Convert base s3 path for sentinel-2 image product into
link to .tif file for true-color-image (TCI) or scene
classification (SCL). <image_type> must be either TCI or SCL"""
s3url_base = s3path.removeprefix("s3://sentinel-cogs")
s3url_base = "https://sentinel-cogs.s3.us-west-2.amazonaws.com" \
+ s3url_base
if image_type == "SCL":
return s3url_base + "/SCL.tif"
elif image_type == "TCI":
return s3url_base + "/TCI.tif"
else:
raise ValueError(
f"Image type must be 'TCI' or 'SCL', Got {image_type} instead")
def get_precise_cloud_percentage(s3_scl_url: str,
shape: Polygon,
) -> float:
""" Get percentage of cloud or cloud shadow
pixels in the Scene Classification at <s3_scl_url>
cropped to <shape> """
with rasterio.open(s3_scl_url) as src:
out_image, _ = mask(src, [shape], crop=True)
# these values represent errors, clouds, or cloud shadows
# see https://sentinels.copernicus.eu/web/sentinel/technical-guides/sentinel-2-msi/level-2a/algorithm-overview#classification-mask-generation
bad_vals = [1, 3, 8, 9, 10]
bad_vals_count = np.bincount(out_image.flatten(),
minlength=11)[bad_vals].sum()
bad_vals_perc = bad_vals_count/np.count_nonzero(out_image)*100
return bad_vals_perc
def check_clouds_crop_save(s3path: str, raster_proj_epsg: int,
save_folder: int, shape: Polygon,
max_cloud_cover_perc: int,
shape_proj_epsg=4326,
) -> Tuple[str, np.ndarray]:
""" Download part of raster image at <s3path> overlapping <shape>,
checks whether overlapping area has less than <max_cloud_cover_perc> clouds,
and if so saves the cropped raster to <save_folder>.
Returns path of saved raster"""
# 1. project shape into same CRS as raster image
shape_projected = project_shape(shape, shape_proj_epsg,
raster_proj_epsg)
# 2. check if the image masked to the shape contains clouds
# by analyzing scene classification file (SCL)
s3url_scl = s3path_to_tif_download_link(s3path, "SCL")
try:
cloud_perc = get_precise_cloud_percentage(s3url_scl, shape_projected)
if (cloud_perc > max_cloud_cover_perc):
print(f"{s3path}: Too many clouds over area of interest ({cloud_perc:.2f}%)")
return None, None
# rasterio throws ValueError if image does not intersect with shape
# this happens if bounding box used in query overlaps with image,
# but not the actual shape
except ValueError:
print(f"{s3path}: Area of interest and image do not overlap")
return None, None
# 3. download true-color image (TCI) overlapping shape into memory
with rasterio.open(s3path_to_tif_download_link(s3path, "TCI")) as src:
out_image, out_transform = mask(src, [shape_projected],
crop=True)
out_meta = src.meta
# 4. write cropped image to file
os.makedirs(save_folder, exist_ok=True)
out_meta.update({"driver": "GTiff",
"height": out_image.shape[1],
"width": out_image.shape[2],
"transform": out_transform})
out_path = save_folder + s3path.split("/")[-1] + "-TCI_cropped.tif"
with rasterio.open(out_path, "w", **out_meta) as dest:
dest.write(out_image)
print(f"{s3path}: Wrote image to {out_path}")
return out_path
image_metadata_df = image_metadata_df.head(10)
# save the cropped images into folders grouped by areas of interest
name_to_folder = lambda name: "./data/" + name + "/"
image_metadata_df["file"] = None
image_metadata_df["file"] = image_metadata_df.apply(
lambda row: check_clouds_crop_save(
s3path=row["earthsearch:s3_path"],
raster_proj_epsg=row["proj:epsg"],
shape=row["area_geom"],
save_folder=name_to_folder(row["area_name"]),
max_cloud_cover_perc=MAX_CLOUD_COVER_PRECISE),
axis=1)
downloaded_images = image_metadata_df[image_metadata_df.file.notna()]
downloaded_images.to_csv("./data/images_overview.csv", index=False)
downloaded_images[["area_name", "datetime"]].sample(3).head()And we are done!
You can find a notebook with all the code from this post in my Github here.
If someone reading has some experience with better ways of approaching any of these steps (which I am sure exist!), I would love to hear about it.